
中国农学通报 ›› 2021, Vol. 37 ›› Issue (23): 29-37.doi: 10.11924/j.issn.1000-6850.casb2020-0837
所属专题: 生物技术
石会玲1,2( ), 周宇航1,2, 何平1,2, 黄蒙蒙1,2, 邵帅1,2, 葛菁萍1,2, 凌宏志1,2(
), 周宇航1,2, 何平1,2, 黄蒙蒙1,2, 邵帅1,2, 葛菁萍1,2, 凌宏志1,2( )
)
收稿日期:2020-12-28
修回日期:2021-04-13
出版日期:2021-08-15
发布日期:2021-08-26
通讯作者:
凌宏志
作者简介:石会玲,女,1997年出生,黑龙江穆棱人,硕士研究生,研究方向:微生物资源挖掘与利用。通信地址:150080 黑龙江哈尔滨南岗区学府路74号 黑龙江大学生命科学学院,E-mail:
Shi Huiling1,2( ), Zhou Yuhang1,2, He Ping1,2, Huang Mengmeng1,2, Shao Shuai1,2, Ge Jingping1,2, Ling Hongzhi1,2(
), Zhou Yuhang1,2, He Ping1,2, Huang Mengmeng1,2, Shao Shuai1,2, Ge Jingping1,2, Ling Hongzhi1,2( )
)
Received:2020-12-28
Revised:2021-04-13
Online:2021-08-15
Published:2021-08-26
Contact:
Ling Hongzhi
摘要:
旨在应用自杀质粒重组技术,构建阴沟肠杆菌乳酸脱氢酶突变株,为进一步提高乙偶姻的产量和扩大菌株选择范围奠定基础。利用双酶切的方法将同源片段插入到自杀质粒pKR6K中,构建出ldh基因敲除质粒,然后利用细菌接合的方法敲除E. cloacae的ldh基因。成功克隆出两段E. cloacae乳酸脱氢酶基因的同源序列,长度分别为526 bp,通过序列比对分析,E. cloacae乳酸脱氢酶基因序列相似性为100%。通过对E. cloacae进行乳酸脱氢酶基因的敲除,成功构建一株ldh缺失重组菌株E. cloacae△ldh,同时2,3-丁二醇提高6.8%,乙酸提高了24.3%。E. cloacae乳酸脱氢酶缺失工程菌株构建成功,对利用微生物法工业化生产乙偶姻奠定基础。
中图分类号:
石会玲, 周宇航, 何平, 黄蒙蒙, 邵帅, 葛菁萍, 凌宏志. 阴沟肠杆菌乳酸脱氢酶基因缺失突变株的构建及其生物学特性[J]. 中国农学通报, 2021, 37(23): 29-37.
Shi Huiling, Zhou Yuhang, He Ping, Huang Mengmeng, Shao Shuai, Ge Jingping, Ling Hongzhi. Lactic Dehydrogenase Gene Deletion Mutant of Enterobacter cloacae: Construction and Biological Characteristics[J]. Chinese Agricultural Science Bulletin, 2021, 37(23): 29-37.
| 引物 | 序列5’-3’ | 酶切位点 | 用途 |
|---|---|---|---|
| ldh1F | AATTxxxxxGAATTChhhhhACCGTGTTAAGTTCAAGCGCACCAA | EcoRI | 克隆ldh基因上游片段526 bp |
| ldh1R | AATTxxxxxGAATTCGGATCChhhhhAAGACTTTCTCCAGTGATTTTACAT | EcoRI, BamHI | |
| ldh2F | AATTxxxxxTCTAGAhhhhhGCCGACATGCCGGGTGGCGGTTACG | XbaI | 克隆ldh基因下游片段526 bp |
| ldh2R | AATTxxxxxGCATGCGTCGAChhhhhGGCGACGGTCATTATTTCGCAGGCG | SphI, SalI | |
| ldh-up | TTTTTGGCGCAACGGTTGACGGTGC | — | 验证ldh基因敲除结果 |
| ldh-down | ATGCGGGTCGCCGCCGCGCCTGCCA | — | |
| ldhF | CGGCTTAGACTATCTCGTTAGGACAC | — | 克隆ldh基因 |
| ldhR | GTCTTATGAAACTCGCGGTATATAGCAC | — |
| 引物 | 序列5’-3’ | 酶切位点 | 用途 |
|---|---|---|---|
| ldh1F | AATTxxxxxGAATTChhhhhACCGTGTTAAGTTCAAGCGCACCAA | EcoRI | 克隆ldh基因上游片段526 bp |
| ldh1R | AATTxxxxxGAATTCGGATCChhhhhAAGACTTTCTCCAGTGATTTTACAT | EcoRI, BamHI | |
| ldh2F | AATTxxxxxTCTAGAhhhhhGCCGACATGCCGGGTGGCGGTTACG | XbaI | 克隆ldh基因下游片段526 bp |
| ldh2R | AATTxxxxxGCATGCGTCGAChhhhhGGCGACGGTCATTATTTCGCAGGCG | SphI, SalI | |
| ldh-up | TTTTTGGCGCAACGGTTGACGGTGC | — | 验证ldh基因敲除结果 |
| ldh-down | ATGCGGGTCGCCGCCGCGCCTGCCA | — | |
| ldhF | CGGCTTAGACTATCTCGTTAGGACAC | — | 克隆ldh基因 |
| ldhR | GTCTTATGAAACTCGCGGTATATAGCAC | — |
| PCR反应体系组分 | 添加量/μL | 终浓度 |
|---|---|---|
| Template DNA | 1 | — |
| Forward primer (10 μmol/L) | 1 | 0.2 μmol/L |
| Reverse primer (10 μmol/L) | 1 | 0.2 μmol/L |
| TransStart® FastPfu DNA Polymerase | 1 | 2.5 units |
| 5× TransStart® FastPfu Buffer | 10 | 1× |
| dNTPs (2.5 mmol/L) | 4 | 0.2 mmol/L |
| ddH2O | Up to 50 | — |
| PCR反应体系组分 | 添加量/μL | 终浓度 |
|---|---|---|
| Template DNA | 1 | — |
| Forward primer (10 μmol/L) | 1 | 0.2 μmol/L |
| Reverse primer (10 μmol/L) | 1 | 0.2 μmol/L |
| TransStart® FastPfu DNA Polymerase | 1 | 2.5 units |
| 5× TransStart® FastPfu Buffer | 10 | 1× |
| dNTPs (2.5 mmol/L) | 4 | 0.2 mmol/L |
| ddH2O | Up to 50 | — |
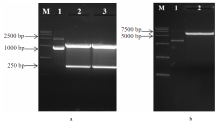
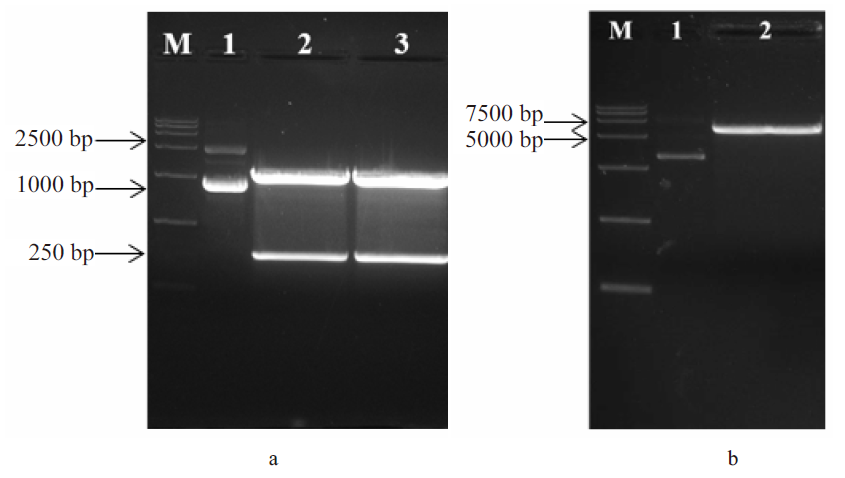
| 产物浓度/(g/L) | 菌株 | 变化情况 | |
|---|---|---|---|
| E. cloacae | E. cloacae△ldh | ||
| 乙偶姻 | 2.83±0.48a(48 h) | 3.05±0.27a(48 h) | — |
| 乳酸 | 2.85±0.21a(12 h) | 0.01±0.01b(48h) | ↓ |
| 2,3-BD | 17.11±0.51b(12 h) | 18.28±0.42a(12 h) | ↑ |
| 丁二酸 | 2.08±0.24b(48 h) | 2.46±0.10a(24 h) | ↑ |
| 乙酸 | 2.92±0.20b(48 h) | 3.63±0.31a(48 h) | ↑ |
| 乙醇 | 2.81±0.11a(24 h) | 3.17±0.31a(24 h) | — |
| 产物浓度/(g/L) | 菌株 | 变化情况 | |
|---|---|---|---|
| E. cloacae | E. cloacae△ldh | ||
| 乙偶姻 | 2.83±0.48a(48 h) | 3.05±0.27a(48 h) | — |
| 乳酸 | 2.85±0.21a(12 h) | 0.01±0.01b(48h) | ↓ |
| 2,3-BD | 17.11±0.51b(12 h) | 18.28±0.42a(12 h) | ↑ |
| 丁二酸 | 2.08±0.24b(48 h) | 2.46±0.10a(24 h) | ↑ |
| 乙酸 | 2.92±0.20b(48 h) | 3.63±0.31a(48 h) | ↑ |
| 乙醇 | 2.81±0.11a(24 h) | 3.17±0.31a(24 h) | — |
| [1] | Xiao Z, Lu J R. Generation of Acetoin and Its Derivatives in Foods[J]. Journal of Agricultural & Food Chemistry, 2014, 62(28):6487-97. |
| [2] |
Xiao Z, Xu P. Acetoin metabolism in bacteria[J]. Critical Reviews in Microbiology, 2007, 33(2):127-140.
doi: 10.1080/10408410701364604 URL |
| [3] | 刘晓霏, 付晶, 霍广鑫, 等. 生物法制备平台化合物乙偶姻的最新研究进展[J]. 中国生物工程杂志, 2015, 35(10):91-99. |
| [4] | 张小舟, 曾崇余, 任晓乾. 乙偶姻合成工艺[J]. 南京化工大学学报:自然科学版, 2001. |
| [5] | 胡明一, 王中. 食用香料乙偶姻[J]. 精细与专用化学品, 2002, 10(1):20-21. |
| [6] |
Odile M, M B, Pascal L, Patrick A D. GRIMONT. Taxonomic Diversity of the D-Glucose Oxidation Pathway in the Enterobacteriaceae[J]. International Journal of Systematic Bacteriology, 1989, 39(1):61-67.
doi: 10.1099/00207713-39-1-61 URL |
| [7] | 葛岚, 邵晓丛, 吴晓敏, 等. 工业化制备2,3-丁二醇的新途径[J]. 科技创新导报, 2009(33):106. |
| [8] |
Choi E J, Kim J W, Kim S J, et al. Enhanced production of 2,3-butanediol in pyruvate decarboxylase-deficient Saccharomyces cerevisiae through optimizing ratio of glucose/galactose[J]. Biotechnol J, 2016, 11(11):1424-1432.
doi: 10.1002/biot.v11.11 URL |
| [9] | Stefano R, Davide P, Giulio Z, et al. Effect of oxygen mass transfer rate on the production of 2,3-butanediol from glucose and agro-industrial byproducts by Bacillus licheniformis ATCC9789[J]. Biotechnology for Biofuels, 2018, 11(1). |
| [10] | Kim D K, Rathnasingh C, Song H, et al. Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2,3-butanediol production[J]. Journal of Bioscience & Bioengineering, 2013, 116(2):186-192. |
| [11] | Birajdar S D, Rajagopalan S, Sawant J S, et al. Continuous predispersed solvent extraction process for the downstream separation of 2,3-butanediol from fermentation broth[J]. Separation & Purification Technology, 2015, 151:115-123. |
| [12] |
Ji X J, Liu L G, Shen M Q, et al. Constructing a synthetic metabolic pathway inEscherichia colito produce the enantiomerically pure (R, R)-2,3-butanediol[J]. Biotechnology and Bioengineering, 2015, 112(5):1056-1059.
doi: 10.1002/bit.v112.5 URL |
| [13] | Tong Y J, Ji X J, Shen M Q, et al. Constructing a synthetic constitutive metabolic pathway in Escherichia coli for (R, R)-2,3-butanediol production[J]. Applied Microbiology & Biotechnology, 2016, 100(2). |
| [14] |
Xu Y, Chu H, Gao C, et al. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol[J]. Metabolic Engineering, 2014, 23(5):22-33.
doi: 10.1016/j.ymben.2014.02.004 URL |
| [15] |
Yang Z, Zhang Z. Production of (2R,3R)-2,3-butanediol using engineered Pichia pastoris : strain construction, characterization and fermentation[J]. Biotechnology for Biofuels, 2018, 11(1):35.
doi: 10.1186/s13068-018-1031-1 URL |
| [16] |
Gao Y, Huang H, Chen S, et al. Production of optically pure 2,3-butanediol from Miscanthus floridulus hydrolysate using engineered Bacillus licheniformis strains[J]. World Journal of Microbiology & Biotechnology, 2018, 34(5):66.
doi: 10.1007/s11274-018-2450-7 URL |
| [17] | 王金星. B29菌株LPS合成基因缺失突变株的构建及分析[D]. 上海:上海交通大学, 2014. |
| [18] | He Y X, Hui X U, Fei Y E, et al. Constuction of suiside vector of aroA gene of Haemophilus parasuis[J]. Heilongjiang Animal Science and Veterinary Medicine, 2011(7):20-22. |
| [19] | 于慧敏, 马玉超. 工业微生物代谢途径调控的基因敲除策略[J]. 生物工程学报, 2010, 26(9):1199-1208. |
| [20] | 戴旭明, 薛红, 杨桦, 等. 基因打靶置换型载体的构建和应用研究[J]. 第二军医大学学报, 1998, 19(1):5-8. |
| [21] | 王鸿姣. 基因敲除技术[J]. 农村科学实验, 2017(4). |
| [22] | Xiao Z J, Liu P H, Qin J Y, et al. Statistical optimization of medium components for enhanced acetoin production from molasses and soybean meal hydrolysate[J]. Applied Microbiology & Biotechnology, 2007, 74(1):61-68. |
| [23] | Bornstein N, Fleurette J. Acetoin Production in the Identification of Isolates as Members of Staphylococcus intermedius Hájek[J]. International Journal of Systematic Bacteriology, 1981, 31(3). |
| [24] | Zhang L, Liu Q, Ge Y, et al. Biotechnological production of acetoin, a bio-based platform chemical, from a lignocellulosic resource by metabolically engineered Enterobacter cloacae[J]. Green Chemistry, 2016, 18. |
| [25] | Hillman J D, Andrews S W, Dzuback A L. Acetoin production by wild-type strains and a lactate dehydrogenase-deficient mutant of Streptococcus mutans[J]. Infection & Immunity, 1987, 55(6):1399-1402. |
| [26] | Liu D, Chen Y, Ding F, et al. Simultaneous production of butanol and acetoin by metabolically engineered Clostridium acetobutylicum[J]. Metabolic Engineering, 2015. 27 |
| [27] | 饶志明, 包腾, 张显, 等. 加强表达枯草芽孢杆菌葡萄糖-6-磷酸脱氢酶提高乙偶姻产量[P]. 2015. |
| [28] |
Xu Q M, Xie L X, Li Y Y, et al. Metabolic engineering of Escherichia coli for efficient production of (3R)-acetoin[EB/OJ]. Journal of Chemical Technology & Biotechnology, 2014.DOI 10.1002/jctb.4293.
doi: 10.1002/jctb.4293 |
| [1] | 高慧颖, 姜帆. 龙眼干制前后缓解小鼠疲劳作用的研究[J]. 中国农学通报, 2022, 38(11): 106-110. |
| [2] | 康杰, 王长丽, 葛菁萍. 单倍体酿酒酵母的丙酮酸脱羧酶基因(pdc1)的敲除与鉴定[J]. 中国农学通报, 2020, 36(24): 91-98. |
| [3] | 谢金东,杨燕燕,林 玮,俞春英,周建华,刘德强,王训立. 基于子代基因型鉴定技术研究FMR1敲除 对C57BL/6小鼠繁殖性能的影响[J]. 中国农学通报, 2015, 31(11): 68-71. |
| [4] | 林海龙 付畅 郑副帅 任南琪. 阴沟肠杆菌flavocytochrome C基因克隆及序列分析[J]. 中国农学通报, 2011, 27(18): 228-232. |
| [5] | 林海龙 李伟光 任南琪. 阴沟肠杆菌的Hy3AL和hycA部分基因克隆及序列分析[J]. 中国农学通报, 2011, 27(15): 233-238. |
| [6] | 化占勇,刘雅莉,徐伟荣,王跃进,张雄飞. 同源重组快速构建百合LCHS2基因RNAi表达载体及其鉴定[J]. 中国农学通报, 2010, 26(2): 38-44. |
| [7] | 徐运杰,方热军. 培养液磷含量和pH对鸡离体小肠磷吸收的影响[J]. 中国农学通报, 2009, 25(7): 1-6. |
| [8] | 卢从德 王代钢 杨金宏 禚苏. 高效表达dsRNA大肠杆菌BL21 RNase Ⅲ- 的构建[J]. 中国农学通报, 2009, 25(22): 0-0. |
| [9] | 张冬梅,石振华,林歧,鲁国东,王宗华. 一个假定的稻瘟病菌RhoGEF蛋白参与营养生长和产孢过程的调控[J]. 中国农学通报, 2009, 25(11): 161-164. |
| [10] | 林抗美,官雪芳,马丽娜,王宗华,鲁国东,刘 波. 有机磷农药降解菌—阴沟肠杆菌的生物学特性[J]. 中国农学通报, 2008, 24(9): 382-386. |
| [11] | 马丽娜,官雪芳,朱育菁,林抗美,刘 波. 乐果降解菌的分离、筛选和鉴定[J]. 中国农学通报, 2008, 24(7): 441-444. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||