
中国农学通报 ›› 2020, Vol. 36 ›› Issue (29): 69-77.doi: 10.11924/j.issn.1000-6850.casb2020-0127
所属专题: 生物技术
刘磊1,2( ), 李娜1,2, 姜雪雍1,2, 孙健1,2, 吕雨泽1,2, 葛菁萍1,2(
), 李娜1,2, 姜雪雍1,2, 孙健1,2, 吕雨泽1,2, 葛菁萍1,2( )
)
收稿日期:2020-05-25
修回日期:2020-07-27
出版日期:2020-10-15
发布日期:2020-10-16
通讯作者:
葛菁萍
作者简介:刘磊,男,1996年出生,山东菏泽人,硕士,研究方向:微生物资源挖掘与利用。通信地址:150080 黑龙江省哈尔滨市南岗区学府路74号224信箱 黑龙江大学生命科学学院,Tel:0451-86609016,E-mail:基金资助:
Liu Lei1,2( ), Li Na1,2, Jiang Xueyong1,2, Sun Jian1,2, Lv Yuze1,2, Ge Jingping1,2(
), Li Na1,2, Jiang Xueyong1,2, Sun Jian1,2, Lv Yuze1,2, Ge Jingping1,2( )
)
Received:2020-05-25
Revised:2020-07-27
Online:2020-10-15
Published:2020-10-16
Contact:
Ge Jingping
摘要:
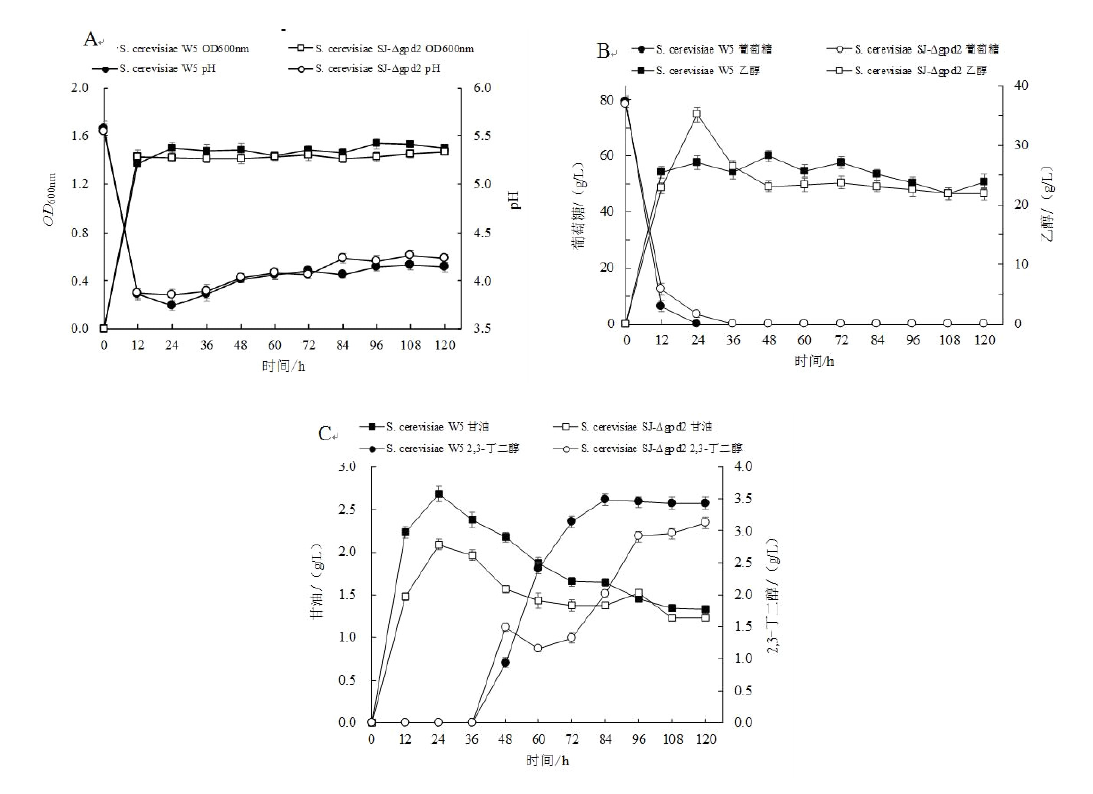
旨在利用CRISPR/Cas9基因编辑技术敲除酿酒酵母甘油-3-磷酸脱氢酶基因(gpd2),探究其对2,3-丁二醇产量的影响。根据酿酒酵母(Saccharomyces cerevisiae)W5甘油-3-磷酸脱氢酶基因(gpd2)设计供体片段及gRNA片段,将gRNA片段与可表达Cas9蛋白的敲除载体相连,之后将重组质粒及供体DNA片段转化到S. cerevisiae W5细胞中,根据表型筛选及PCR验证获得gpd2基因缺失菌株。结果表明目的基因gpd2敲除成功,基因缺失菌株与原始菌株经发酵实验相比,甘油产量下降22.01%,乙醇产量提高24.65%,2,3-丁二醇产量下降10.60%。gpd2基因的敲除并没有提高2,3-丁二醇的产量,原因可能是逐渐积累的NADH会优先被细胞内大量的乙醇脱氢酶所氧化,作用于乙醇的产生,而不是优先作用于2,3-丁二醇的合成。本实验构建了适用于酿酒酵母的基因敲除系统,该系统对进一步探究酿酒酵母其他代谢产物与2,3-丁二醇合成之间的关系具有实际的借鉴意义。
中图分类号:
刘磊, 李娜, 姜雪雍, 孙健, 吕雨泽, 葛菁萍. CRISPR/Cas9技术敲除酿酒酵母gpd2基因对产2,3-丁二醇的影响[J]. 中国农学通报, 2020, 36(29): 69-77.
Liu Lei, Li Na, Jiang Xueyong, Sun Jian, Lv Yuze, Ge Jingping. Effects on 2,3-butanediol Production of Saccharomyces cerevisiae: gpd2 Gene Knockout by CRISPR/Cas9 Technology[J]. Chinese Agricultural Science Bulletin, 2020, 36(29): 69-77.
| 质粒名称 | 质粒特征 | 来源 |
|---|---|---|
| pUDP004 | amdS、TEF1prom-Cas9-PHO5term、panARSopt、GAP prom-CYC1term | 购自ADDGENE |
| pUDP004-gpd2 | pUDP004、GAP prom-sgRNA(gpd2)-CYC1 term | 本实验室构建 |
| pMD18-T | AmpR | 购自宝生物工程(大连)有限公司 |
| 质粒名称 | 质粒特征 | 来源 |
|---|---|---|
| pUDP004 | amdS、TEF1prom-Cas9-PHO5term、panARSopt、GAP prom-CYC1term | 购自ADDGENE |
| pUDP004-gpd2 | pUDP004、GAP prom-sgRNA(gpd2)-CYC1 term | 本实验室构建 |
| pMD18-T | AmpR | 购自宝生物工程(大连)有限公司 |
| 引物名称 | 引物序列(5′→3′) | 引物长度/bp |
|---|---|---|
| GPD2-6F | CAAAAAGATC | 10 |
| GPD2-6R | TCAGGATCTT | 10 |
| GPD2-20F | GATCTTTTACACTCCATCAAGT | 22 |
| GPD2-20R | TTGATGGAGTGTAAAAGATCGA | 22 |
| GPD2-ORF-F | ATGCTTGCTGTCAGAAGATTAACAAGATACACATTCC | 37 |
| GPD2-ORF-R | CTATTCGTCATCGATGTCTAGCTCTTCAATCATCTC | 36 |
| GPD2-F1 | CAGACGCAGCAGCAAGTAAC | 20 |
| GPD2-R1 | TTCGTACACAGCGTTGACCT | 20 |
| GPD2-F2 | AAAGAGGCAAGGGGAGCGAAGGAAAAGGA | 29 |
| GPD2-R2 | TCGCTCCCCTTGCCTCTTTTTCCCCCAACCA | 31 |
| GPD2-300F | GATGGGTTGCTGAGGGGAAG | 20 |
| GPD2-300R | ACTGGAGAGCCGTCAGTAGT | 20 |
| HH-BbsI-F | GCAAATCGTCTTCACCTGAAGACTG | 25 |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC | 24 |
| M13R-48 | AGCGGATAACAATTTCACACAGGA | 24 |
| GPD2-P1 | GCTCGTCGATCTTTTACACTCCATCAA | 27 |
| GPDpro-F | CGGTAGGTATTGATTGTAATTCTG | 24 |
| CYC1-R | GCGTGAATGTAAGCGTGAC | 19 |
| 引物名称 | 引物序列(5′→3′) | 引物长度/bp |
|---|---|---|
| GPD2-6F | CAAAAAGATC | 10 |
| GPD2-6R | TCAGGATCTT | 10 |
| GPD2-20F | GATCTTTTACACTCCATCAAGT | 22 |
| GPD2-20R | TTGATGGAGTGTAAAAGATCGA | 22 |
| GPD2-ORF-F | ATGCTTGCTGTCAGAAGATTAACAAGATACACATTCC | 37 |
| GPD2-ORF-R | CTATTCGTCATCGATGTCTAGCTCTTCAATCATCTC | 36 |
| GPD2-F1 | CAGACGCAGCAGCAAGTAAC | 20 |
| GPD2-R1 | TTCGTACACAGCGTTGACCT | 20 |
| GPD2-F2 | AAAGAGGCAAGGGGAGCGAAGGAAAAGGA | 29 |
| GPD2-R2 | TCGCTCCCCTTGCCTCTTTTTCCCCCAACCA | 31 |
| GPD2-300F | GATGGGTTGCTGAGGGGAAG | 20 |
| GPD2-300R | ACTGGAGAGCCGTCAGTAGT | 20 |
| HH-BbsI-F | GCAAATCGTCTTCACCTGAAGACTG | 25 |
| M13F-47 | CGCCAGGGTTTTCCCAGTCACGAC | 24 |
| M13R-48 | AGCGGATAACAATTTCACACAGGA | 24 |
| GPD2-P1 | GCTCGTCGATCTTTTACACTCCATCAA | 27 |
| GPDpro-F | CGGTAGGTATTGATTGTAATTCTG | 24 |
| CYC1-R | GCGTGAATGTAAGCGTGAC | 19 |

| [1] |
Białkowska A M. Strategies for efficient and economical 2, 3-butanediol production: new trends in this field[J]. World Journal of Microbiology and Biotechnology, 2016,32(12):200-213.
URL pmid: 27778222 |
| [2] | Pasaye Anaya L, Vargas Tah A, Martínez Cámara C, et al. Production of 2, 3-butanediol by fermentation of enzymatic hydrolysed bagasse from agave mezcal‐waste using the native Klebsiella oxytoca UM2-17 strain[J]. Journal of Chemical Technology and Biotechnology, 2019,94(12):3915-3923. |
| [3] |
Shi L T, Gao S S, Yu Y, et al. Microbial production of 2, 3-butanediol by a newly-isolated strain of Serratia marcescens[J]. Biotechnology Letters, 2014,36(5):969-973.
doi: 10.1007/s10529-013-1433-x URL pmid: 24375234 |
| [4] | Thapa L P, Lee S J, Park C, et al. Metabolic engineering of Enterobacter aerogenes to improve the production of 2, 3-butanediol[J]. Biochemical Engineering Journal, 2019,143:169-178. |
| [5] | Lee Y G, Seo J H. Production of 2,3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae[J]. Biotechnology for biofuels, 2019,12(1):204-216. |
| [6] | Ng C, Jung M Y, Lee J, et al. Production of 2, 3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering[J]. Microbial Cell Factories, 2012,11(1):68-81. |
| [7] |
Kim J W, Kim J, Seo S O, et al. Enhanced production of 2, 3-butanediol by engineered Saccharomyces cerevisiae through fine-tuning of pyruvate decarboxylase and NADH oxidase activities[J]. Biotechnology for biofuels, 2016,9:265-276.
URL pmid: 27990176 |
| [8] |
Klein M, Swinnen S, Thevelein J M, et al. Glycerol metabolism and transport in yeast and fungi: established knowledge and ambiguities[J]. Environmental Microbiology, 2017,19(3):878-893.
doi: 10.1111/1462-2920.13617 URL pmid: 27878932 |
| [9] |
Albertyn J, Hohmann S, Thevelein J M, et al. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway[J]. Molecular and Cellular Biology, 1994,14(6):4135-4144.
URL pmid: 8196651 |
| [10] |
Nissen T L, Hamann C W, Kielland-Brandt M C, et al. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synjournal[J]. Yeast, 2000,16(5):463-474.
URL pmid: 10705374 |
| [11] |
Björkqvist S, Ansell R, Adler L, et al. Physiological response to anaerobicity of glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae[J]. Applied and Environmental Microbiology, 1997,63(1):128-132.
URL pmid: 8979347 |
| [12] |
Horwitz A A, Walter J M, Schubert M G, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas[J]. Cell systems, 2015,1(1):88-96.
doi: 10.1016/j.cels.2015.02.001 URL pmid: 27135688 |
| [13] |
Jakočiūnas T, Jensen M K, Keasling J D. CRISPR/Cas9 advances engineering of microbial cell factories[J]. Metabolic Engineering, 2016,34:44-59.
doi: 10.1016/j.ymben.2015.12.003 URL pmid: 26707540 |
| [14] |
Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012,337(6096):816-821.
doi: 10.1126/science.1225829 URL pmid: 22745249 |
| [15] |
Jiang F G, Doudna J A. CRISPR-Cas9 structures and mechanisms[J]. Annual review of biophysics, 2017,46:505-529.
doi: 10.1146/annurev-biophys-062215-010822 URL pmid: 28375731 |
| [16] |
Ge J P, Sun H B, Song G, et al. A genome shuffling-generated Saccharomyces cerevisiae isolate that ferments xylose and glucose to produce high levels of ethanol[J]. Journal of Industrial Microbiology & Biotechnology, 2012,39(5):777-787.
URL pmid: 22270888 |
| [17] | Lee Y G, Seo J H. Production of 2, 3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae[J]. Biotechnology for Biofuels, 2019,12(1):204-216. |
| [18] |
Ehsani M, Fernández M R, Biosca J A, et al. Engineering of 2, 3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae[J]. Applied and Environmental Microbiology, 2009,75(10):3196-3205.
URL pmid: 19329666 |
| [19] |
Ain Q U, Chung J Y, Kim Y H. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN[J]. Journal of Controlled Release, 2015,205:120-127.
doi: 10.1016/j.jconrel.2014.12.036 URL pmid: 25553825 |
| [20] |
de Vries A R G, de Groot P A, van den Broek M, et al. CRISPR-Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus[J]. Microbial Cell Factories, 2017,16(1):222-239.
URL pmid: 29207996 |
| [21] | 刘奎, 梁丽敏, 李振辉, 等. CRISPR/Cas9介导的酿酒酵母ADH2基因中断及反义RNA干扰GPD1的表达[J]. 现代食品科技, 2018,34(10):70-77. |
| [22] |
Shi S B, Liang Y Y, Zhang M M, et al. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae[J]. Metabolic Engineering, 2016,33:19-27.
URL pmid: 26546089 |
| [23] | Liu K, Yuan X, Liang L M, et al. Using CRISPR/Cas9 for multiplex genome engineering to optimize the ethanol metabolic pathway in Saccharomyces cerevisiae[J]. Biochemical Engineering Journal, 2019,145:120-126. |
| [1] | 李建荣, 王伟, 罗定国, 马晓霞, 徐美玲, 滚双宝, 杨巧丽. 合作猪HMOX1基因过表达载体构建及组织表达分析[J]. 中国农学通报, 2022, 38(29): 140-145. |
| [2] | 权莹, 张晓娟, 赵辉, 孙晓敏, 马秀奇. CRISPER/Cas9系统在植物基因组定点修饰及作物遗传育种中的应用研究进展[J]. 中国农学通报, 2022, 38(26): 9-14. |
| [3] | 王长丽, 廖巍, 叶广彬, 葛菁萍, 刘磊, 马毓坚, 黄霞, 宾晓芸. 编码酿酒酵母丙酮酸脱羧酶(Pdc6)基因克隆及其生物信息学分析[J]. 中国农学通报, 2021, 37(9): 103-108. |
| [4] | 高忠奎, 蒋菁, 韩柱强, 黄志鹏, 熊发前, 唐秀梅, 吴海宁, 钟瑞春, 刘菁, 唐荣华, 贺梁琼. CRISPR/Cas9系统及其在粮油作物遗传改良中的研究进展[J]. 中国农学通报, 2021, 37(20): 26-34. |
| [5] | 张弛, 吕雨泽, 邓利廷, 孙健, 葛菁萍. 外源添加乙偶姻对酿酒酵母产2,3-丁二醇及其菌株的影响[J]. 中国农学通报, 2021, 37(2): 20-27. |
| [6] | 王婷甄, 孙燕川, 唐文琨, 范双喜, 郝敬虹. 叶用莴苣无缝克隆构建LsE3基因过表达载体和新RNAi载体及遗传转化体系优化[J]. 中国农学通报, 2021, 37(11): 15-23. |
| [7] | 丁昊, 刘文娟, 孙健, 刘磊, 平文祥, 葛菁萍. 高产2,3-丁二醇的潜在酿酒酵母菌株筛选[J]. 中国农学通报, 2020, 36(24): 107-115. |
| [8] | 康杰, 王长丽, 葛菁萍. 单倍体酿酒酵母的丙酮酸脱羧酶基因(pdc1)的敲除与鉴定[J]. 中国农学通报, 2020, 36(24): 91-98. |
| [9] | 杨智宇, 佟天奇, 刘磊, 平文祥, 葛菁萍. 外源添加乙偶姻对酿酒酵母W5/W141产2,3-丁二醇的影响[J]. 中国农学通报, 2020, 36(23): 19-25. |
| [10] | 杨智宇, 佟天奇, 刘磊, 平文祥, 葛菁萍. 外源添加乙酸对酿酒酵母(Saccharomyces cerevisiae)产2,3-丁二醇影响初探[J]. 中国农学通报, 2020, 36(21): 104-112. |
| [11] | 郑文涌, 杨涛, 李双全, 吕常旭, 石敏, 马立保, 晏向华. 新型酿酒酵母培养物对育肥猪生产性能、肌肉品质和肠道微生物的影响[J]. 中国农学通报, 2020, 36(21): 145-154. |
| [12] | 孙少慧,张静华,杨帅,高云飞,闵凡祥,马 纪,吕典秋,金光辉,王晓丹. 马铃薯晚疫病菌RxLR效应因子RD24基因克隆及其PVX表达载体构建与鉴定[J]. 中国农学通报, 2019, 35(5): 144-149. |
| [13] | 吕晶森,谭成方,刘春林,阮 颖. CRISPR/Cas9技术在获取双突变体材料中的应用[J]. 中国农学通报, 2018, 34(8): 27-36. |
| [14] | 徐 晶,张桂山,孙丽敏,白 曼,项露杰,姜怀志. 辽宁绒山羊皮肤毛囊mir-1298-5p靶基因预测及表达载体构建[J]. 中国农学通报, 2018, 34(5): 123-128. |
| [15] | 佟天奇,裴芳艺,王长丽,孙 健,葛菁萍. 酿酒酵母(Saccharomyces cerevisiae)WBG3菌株发酵特性研究[J]. 中国农学通报, 2018, 34(32): 49-56. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||