
中国农学通报 ›› 2023, Vol. 39 ›› Issue (11): 29-35.doi: 10.11924/j.issn.1000-6850.casb2022-0219
收稿日期:2022-03-30
修回日期:2022-06-15
出版日期:2023-04-15
发布日期:2023-04-10
通讯作者:
李杰,男,1983年出生,湖南常德人,副研究员,博士,主要从事花卉栽培与育种方面的研究。通信地址:510640 广州天河区五山路金颖东一街1号 广东省农业科学院环境园艺研究所,Tel:021-85161185,E-mail:作者简介:叶广英,男,1988年出生,山东聊城人,博士,研究方向:花卉栽培与育种。通信地址:510640 广州天河区五山路金颖东一街1号 广东省农业科学院环境园艺研究所,Tel:021-85161185,E-mail:yeggyy20062009@163.com。
基金资助:
YE Guangying( ), WANG Zaihua, LIU Hailin, LI Jie(
), WANG Zaihua, LIU Hailin, LI Jie( )
)
Received:2022-03-30
Revised:2022-06-15
Online:2023-04-15
Published:2023-04-10
摘要:
旨在从转录组水平解析黄蜀葵体内进行的主要生物进程及黄酮醇化合物合成的分子机制。利用高通量测序PacBio Sequel平台,以黄蜀葵的花、茎和叶的混合样品为材料,使用单分子实时测序技术(SMRT)进行全长转录组测序。测序数据质控后共获得53925条高质量转录本,其中53329个基因被公共数据库注释。共16574个基因注释到138条途径上,其中注释最多的途径为代谢途径(8556个),其次为次生代谢物的生物合成途径(4650个)。有10700个黄蜀葵基因发生了可变剪切事件,存在7种可变剪切类型。预测到3287个转录因子,bHLH 转录因子家族的基因最多。检测到5683个SSR位点,且AAG/CCT类型最多。鉴定出黄酮醇合成相关的基因170个,包含10个关键的糖基转移酶基因,且CHS和F3H基因发生了内含子保留型的可变剪切。本研究揭示了黄蜀葵转录组的整体水平及黄酮醇生物合成的相关基因,为后期遗传改良和功能基因挖掘与利用提供了分子基础。
叶广英, 王再花, 刘海林, 李杰. 药食兼用植物黄蜀葵全长转录组测序及特性分析[J]. 中国农学通报, 2023, 39(11): 29-35.
YE Guangying, WANG Zaihua, LIU Hailin, LI Jie. Medicinal and Edible Abelmoschus manihot: Sequencing and Characteristic Analysis of Full-length Transcriptome[J]. Chinese Agricultural Science Bulletin, 2023, 39(11): 29-35.
| 物种 | 转录本数量 | 百分比/% |
|---|---|---|
| 木槿 Hibiscus syriacus | 29933 | 56.14 |
| 雷蒙德氏棉 Gossypium raimondii | 4105 | 7.70 |
| 海岛棉 Gossypium barbadense | 2818 | 5.29 |
| 树棉 Gossypium arboreum | 2555 | 4.79 |
| 陆地棉 Gossypium hirsutum | 2546 | 4.77 |
| 榴莲 Durio zibethinus | 1765 | 3.31 |
| 黄褐棉 Gossypium mustelinum | 1625 | 3.05 |
| 澳洲棉 Gossypium australe | 1570 | 2.94 |
| 夏威夷棉 Gossypium tomentosum | 1264 | 2.37 |
| 可可 Theobroma cacao | 1254 | 2.35 |
| 黄秋葵 Abelmoschus esculentus | 224 | 0.42 |
| 其他 | 3661 | 6.87 |
| 总计 | 53320 | 100 |
| 物种 | 转录本数量 | 百分比/% |
|---|---|---|
| 木槿 Hibiscus syriacus | 29933 | 56.14 |
| 雷蒙德氏棉 Gossypium raimondii | 4105 | 7.70 |
| 海岛棉 Gossypium barbadense | 2818 | 5.29 |
| 树棉 Gossypium arboreum | 2555 | 4.79 |
| 陆地棉 Gossypium hirsutum | 2546 | 4.77 |
| 榴莲 Durio zibethinus | 1765 | 3.31 |
| 黄褐棉 Gossypium mustelinum | 1625 | 3.05 |
| 澳洲棉 Gossypium australe | 1570 | 2.94 |
| 夏威夷棉 Gossypium tomentosum | 1264 | 2.37 |
| 可可 Theobroma cacao | 1254 | 2.35 |
| 黄秋葵 Abelmoschus esculentus | 224 | 0.42 |
| 其他 | 3661 | 6.87 |
| 总计 | 53320 | 100 |
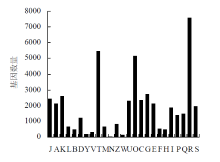

| 参与途径 | 注释基因数量 | 途径代码 |
|---|---|---|
| 代谢途径 | 8556 | ko01100 |
| 次生代谢物的生物合成 | 4650 | ko01110 |
| 碳代谢 | 1670 | ko01200 |
| 氨基酸的生物合成 | 1135 | ko01230 |
| 植物激素信号转导 | 979 | ko04075 |
| 氧化磷酸化 | 916 | ko00190 |
| 内质网中的蛋白质加工 | 848 | ko04141 |
| 核糖体 | 745 | ko03010 |
| 剪接体 | 708 | ko03040 |
| RNA 转运 | 663 | ko03013 |
| 糖酵解/糖质新生 | 657 | ko00010 |
| 淀粉和蔗糖代谢 | 635 | ko00500 |
| 光合作用 | 634 | ko00195 |
| MAPK信号通路-植物 | 620 | ko04016 |
| 内吞作用 | 618 | ko04144 |
| 参与途径 | 注释基因数量 | 途径代码 |
|---|---|---|
| 代谢途径 | 8556 | ko01100 |
| 次生代谢物的生物合成 | 4650 | ko01110 |
| 碳代谢 | 1670 | ko01200 |
| 氨基酸的生物合成 | 1135 | ko01230 |
| 植物激素信号转导 | 979 | ko04075 |
| 氧化磷酸化 | 916 | ko00190 |
| 内质网中的蛋白质加工 | 848 | ko04141 |
| 核糖体 | 745 | ko03010 |
| 剪接体 | 708 | ko03040 |
| RNA 转运 | 663 | ko03013 |
| 糖酵解/糖质新生 | 657 | ko00010 |
| 淀粉和蔗糖代谢 | 635 | ko00500 |
| 光合作用 | 634 | ko00195 |
| MAPK信号通路-植物 | 620 | ko04016 |
| 内吞作用 | 618 | ko04144 |
| 转录因子 | 转录本数量/条 | 百分比/% |
|---|---|---|
| bHLH | 243 | 7.39 |
| TALE | 214 | 6.51 |
| C3H | 183 | 5.57 |
| GRAS | 177 | 5.38 |
| NAC | 176 | 5.35 |
| C2H2 | 175 | 5.32 |
| bZIP | 160 | 4.87 |
| HD-ZIP | 138 | 4.20 |
| MYB_related | 132 | 4.02 |
| ERF | 131 | 3.99 |
| 其他 | 1558 | 47.40 |
| 合计 | 3287 | 100 |
| 转录因子 | 转录本数量/条 | 百分比/% |
|---|---|---|
| bHLH | 243 | 7.39 |
| TALE | 214 | 6.51 |
| C3H | 183 | 5.57 |
| GRAS | 177 | 5.38 |
| NAC | 176 | 5.35 |
| C2H2 | 175 | 5.32 |
| bZIP | 160 | 4.87 |
| HD-ZIP | 138 | 4.20 |
| MYB_related | 132 | 4.02 |
| ERF | 131 | 3.99 |
| 其他 | 1558 | 47.40 |
| 合计 | 3287 | 100 |

| [1] |
doi: 10.3390/molecules23102649 URL |
| [2] |
doi: 10.1016/j.foodchem.2014.08.058 URL |
| [3] |
王雪梅, 施文婷, 吴迷迷, 等. 黄蜀葵胶中的糖分析[J]. 食品科学, 2011, 32:256-260.
doi: 10.7506/spkx1002-6630-201106059 |
| [4] |
doi: 10.1016/j.ijbiomac.2017.08.130 URL |
| [5] |
|
| [6] |
温锐, 谢国勇, 李旭森, 等. 黄蜀葵化学成分与药理活性研究进展[J]. 中国野生植物资源, 2015, 34(2):37-44.
|
| [7] |
刘童童, 王宇阳, 占永立. 基于网络药理学探讨黄葵胶囊治疗IgA肾病的作用机制[J]. 中药新药与临床药理, 2020, 31(9):1079-1085.
|
| [8] |
doi: 10.1002/jssc.v42.11 URL |
| [9] |
doi: 10.1039/C5AY02103K URL |
| [10] |
doi: 10.1007/s13562-021-00687-9 |
| [11] |
doi: 10.1371/journal.pone.0242591 URL |
| [12] |
赵陆滟, 曹绍玉, 许俊强, 等. 全长转录组测序在植物中的应用研究进展[J]. 植物遗传资源学报, 2019, 20(6):1390-1398.
|
| [13] |
孙铭阳, 徐世强, 顾艳, 等. 穿心莲全长转录组测序及特性分析[J]. 中国农学通报, 2021, 37(27):82-89.
doi: 10.11924/j.issn.1000-6850.casb2020-0775 |
| [14] |
doi: 10.1111/tpj.2015.82.issue-6 URL |
| [15] |
doi: 10.1111/tpj.2017.90.issue-1 URL |
| [16] |
王强, 胡金鹏, 潘雅清, 等. 蒙古岩黄芪全长转录组测序及微卫星位点特征分析[J]. 分子植物育种, 2021, 19(24):8109-8120.
|
| [17] |
裴徐梨, 胡文婷, 杨云美, 等. 青花菜全长转录组测序分析[J]. 分子植物育种, 2021, https://kns.cnki.net/kcms/detail/46.1068.S.20210720.1644.008.html.
|
| [18] |
doi: 10.1093/bioinformatics/bti610 pmid: 16081474 |
| [19] |
pmid: 16960968 |
| [20] |
doi: 10.1261/rna.051557.115 pmid: 26179515 |
| [21] |
doi: 10.1038/celldisc.2017.31 URL |
| [22] |
doi: 10.1093/nar/gky066 pmid: 29401301 |
| [23] |
郑乾明, 王小柯, 马玉华. 红肉火龙果成熟茎的全长转录组测序分析及蔗糖代谢相关基因分[J]. 西南农业学报, 2022, 36(3).
|
| [24] |
宋发军, 杨瑞霜, 吕昕芮, 等. 基于PacBio平台的七叶一枝花全长转录组测序[J]. 中南民族大学学报, 2022, 41(2):154-156.
|
| [25] |
梅瑜, 李向荣, 蔡时可, 等. 药食同源植物甘葛藤的全长转录组分析[J]. 华北农学报, 2021, 36(5):10-17.
doi: 10.7668/hbnxb.20192213 |
| [26] |
林颖, 郭秀莲, 朱勋路, 等. 黄蜀葵查尔酮合成酶基因AmCHS克隆及序列分析[J]. 四川大学学报. 2008, 45(6):1327-1352.
|
| [27] |
赵春杰, 李慧慧, 刘德梅, 等. 基于GBS、DArT-array和SSR标记构建普通小麦高密度遗传图谱[J]. 华中农业大学学报, 2019, 38(6):56-61.
|
| [28] |
刘嘉霖, 谢慧敏, 张峥, 等. 基于QTL相关SSR标记分析黄淮海和南方大豆品种的遗传多样性及群体遗传结构[J]. 中国油料作物学报, 2022, 44:63-71.
|
| [29] |
doi: 10.1007/s10722-015-0259-x URL |
| [30] |
刘小红. 基于SMRT 技术的水杉全长转录组分析及基因功能注释[J]. 湖南农业大学学报, 2022, 48(1).
|
| [1] | 付瑜华, 蒙秋伊, 尚昆, 李秀诗, 刘凡值, 李祥栋. 玉米和高粱SSR标记在薏苡中的通用性研究[J]. 中国农学通报, 2023, 39(5): 33-38. |
| [2] | 巩永永, 端木慧子. 甜菜TIFY基因家族的全基因组鉴定与生物信息学分析[J]. 中国农学通报, 2022, 38(8): 17-24. |
| [3] | 余兰, 王浩然, 张莹, 邢红运, 丁琪, 赵宝珍, 崔娜. 转录因子MYCs调控番茄表皮毛萜类化合物的分子机制研究进展[J]. 中国农学通报, 2022, 38(6): 87-93. |
| [4] | 李锐, 尚霄, 尚春树, 常利芳, 闫蕾, 白建荣. SSR荧光检测解析224份山西玉米自交系的遗传结构与遗传关系[J]. 中国农学通报, 2022, 38(5): 9-16. |
| [5] | 郭巨先, 吴廷全, 李沐蓉, 唐康, 姚春鹏, 李桂花, 罗文龙, 王瑞. 基于SSR标记的芋头种质资源遗传多样性分析及抗疫病鉴定[J]. 中国农学通报, 2022, 38(36): 112-119. |
| [6] | 郭栋梁, 黄石连, 王静, 韩冬梅, 李建光. 基于SSR分子标记的龙眼种质资源遗传多样性分析及其指纹图谱构建[J]. 中国农学通报, 2022, 38(36): 67-73. |
| [7] | 徐晓美, 李颖, 衡周, 徐小万, 李涛, 王恒明. 响应辣椒疫霉菌诱导的CaWRKY转录因子筛选及其信号通路分析[J]. 中国农学通报, 2022, 38(32): 22-31. |
| [8] | 赵雅儒, 邳植, 刘蕊, 马语嫣, 吴则东. 不同甜菜单胚细胞质雄性不育系与保持系的遗传多样性分析[J]. 中国农学通报, 2022, 38(30): 35-40. |
| [9] | 马贵芳, 辛海波, 修莉, 孙朝霞, 张华. 荞麦脱壳性状的研究进展[J]. 中国农学通报, 2022, 38(24): 19-27. |
| [10] | 张佳琦, 郭宗珊, 刘长华, 李荣田. 黑龙江省水稻品种的遗传多样性[J]. 中国农学通报, 2022, 38(17): 1-9. |
| [11] | 梁燕, 韩传明, 周继磊, 孙超, 王翠香, 李春明, 王静, 闵旭峰, 公庆党, 孟晓烨, 杨绪强. 山东核桃良种SSR指纹图谱及分子身份证的构建——基于毛细管电泳分析[J]. 中国农学通报, 2022, 38(15): 113-121. |
| [12] | 王晓燕, 单红丽, 张荣跃, 仓晓燕, 王长秘, 李文凤, 尹炯, 罗志明, 黄应昆. 甘蔗抗褐锈病新基因抗感病池构建及SSR多态性引物筛选[J]. 中国农学通报, 2021, 37(6): 97-103. |
| [13] | 马慧敏, 孙培琳, 马春泉. 转录因子BvM14-GAI耐盐功能研究[J]. 中国农学通报, 2021, 37(34): 34-42. |
| [14] | 王雪, 王盛昊, 于冰. 转录因子和启动子互作分析技术及其在植物应答逆境胁迫中的研究进展[J]. 中国农学通报, 2021, 37(33): 112-119. |
| [15] | 孙铭阳, 徐世强, 顾艳, 梅瑜, 周芳, 李静宇, 王继华. 穿心莲全长转录组测序及特性分析[J]. 中国农学通报, 2021, 37(27): 82-89. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||