
中国农学通报 ›› 2022, Vol. 38 ›› Issue (24): 19-27.doi: 10.11924/j.issn.1000-6850.casb2022-0077
马贵芳1,2( ), 辛海波1, 修莉3, 孙朝霞2, 张华1(
), 辛海波1, 修莉3, 孙朝霞2, 张华1( )
)
收稿日期:2022-02-05
修回日期:2022-04-27
出版日期:2022-08-25
发布日期:2022-08-22
通讯作者:
张华
作者简介:马贵芳,女,1996年出生,山西大同人,硕士研究生,研究方向:种质创新与遗传育种。通信地址:100102 北京市朝阳区花家地甲7号 北京市园林绿化科学研究院,E-mail: 基金资助:
MA Guifang1,2( ), XIN Haibo1, XIU Li3, SUN Chaoxia2, ZHANG Hua1(
), XIN Haibo1, XIU Li3, SUN Chaoxia2, ZHANG Hua1( )
)
Received:2022-02-05
Revised:2022-04-27
Online:2022-08-25
Published:2022-08-22
Contact:
ZHANG Hua
摘要:
荞麦适应性广、种质资源丰富,是一种药食同源作物,其籽实具有很高的营养价值。荞麦籽粒由胚、胚乳和外壳构成,外壳富含木质素、纤维素和糖,导致脱壳困难,这一性状严重制约着荞麦加工产业的发展,因此培育易脱壳种质资源是荞麦育种的主要目标之一。荞麦种壳的形成和开裂受遗传和环境共同调控,为了充分了解影响荞麦脱壳的因素,本研究归纳了不同荞麦种壳的组成及细胞排列方式;总结了酶在种子壳裂发生过程中的作用及蜡质角质层在种子外壳脱落过程中的功能;分析了转录因子对壳发育的调控。本研究同时指出了薄壳和厚壳荞麦种壳发育和裂壳性状的主要差异及其关键候选调控基因,为脱壳关键调控基因克隆和高效分子设计育种提供重要的理论依据。
中图分类号:
马贵芳, 辛海波, 修莉, 孙朝霞, 张华. 荞麦脱壳性状的研究进展[J]. 中国农学通报, 2022, 38(24): 19-27.
MA Guifang, XIN Haibo, XIU Li, SUN Chaoxia, ZHANG Hua. Buckwheat Seed Shelling Characters: A Review[J]. Chinese Agricultural Science Bulletin, 2022, 38(24): 19-27.
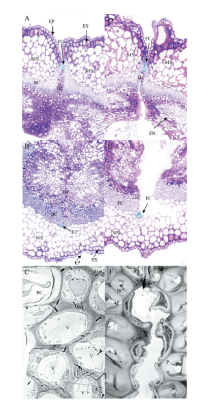
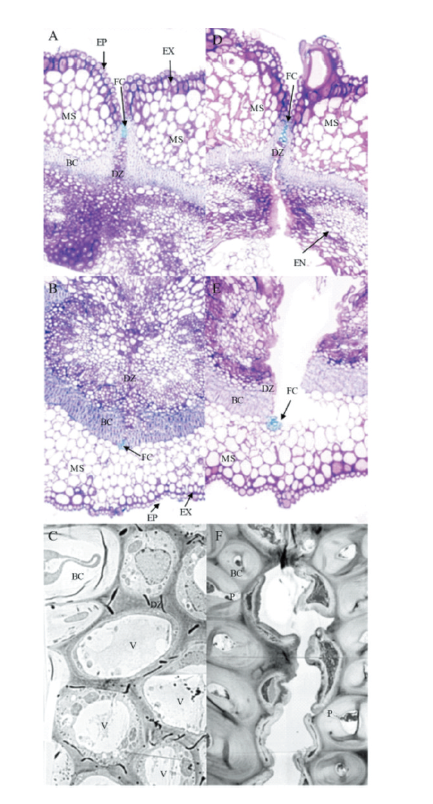
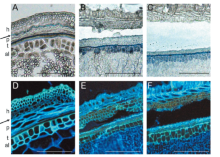
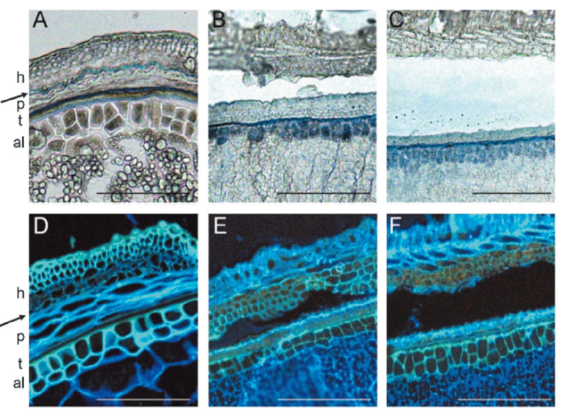
| [1] |
ZHANG L X, LI B, MA Q, et al. The Tartarybuckwheat genome provides insights into rutinbiosynthesis and abiotic stress tolerance[J]. Molecular plant, 2017, 10:1224-1237.
doi: 10.1016/j.molp.2017.08.013 URL |
| [2] |
CHRISTA K, SORALŚMIETANA M. Buckwheat grains and buckwheat products nutritional and prophylactic value of their components-a review[J]. Czech journal of food sciences, 2008, 26:153-162.
doi: 10.17221/1602-CJFS URL |
| [3] | ZHAO G, TANG Y, WANG A H. Research of composition and function of Tartary buckwheat and its development and application[J]. Journal of sichuan agricultural university, 2001, 19:355-358. |
| [4] |
GAO F, ZHOU J, DENG R Y, et al. Overexpression of a Tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis[J]. Journal of plant physiology, 2017, 214:81-90.
doi: 10.1016/j.jplph.2017.04.007 URL |
| [5] |
SUN Z X, LING H B, HOU S Y, et al. Tartary buckwheat FtMYB31 gene encoding an R2R3-MYB transcription factor enhances flavonoid accumulation in tobacco[J]. Journal of plant growth regulation, 2020, 39:564-574.
doi: 10.1007/s00344-019-10000-7 URL |
| [6] | FAOSTAT WWW Document. URL http://www.fao.org/faostat/en/#home, 2019 (accessed 4.15.19). |
| [7] | SARWAR I, THANUSHREE M P, SUDHA M L, et al. Quality characteristics of buckwheat (Fagopyrum esculentum) based nutritious ready-to-eat extruded baked snack[J]. Journal of food science and technology, 2021, 58(5):1-7. |
| [8] |
PODOLSKA G, GUISKA E, KLEPACKA J, et al. Bioactive compounds in different buckwheat species[J]. Plants, 2021, 10(5):961-961.
doi: 10.3390/plants10050961 URL |
| [9] | 于丽萍. 荞麦米的加工[J]. 西部粮油科技, 2002, 27(5):43-44. |
| [10] | 张永林, 易启伟, 王旺平, 等. 立式精碾米机机组的设计[J]. 农业机械学报, 2006, 37(10):222-223. |
| [11] | 陈伟, 杜文亮, 刘彩霞. 苦荞麦浸蒸处理前后的力学特性[J]. 农机化研究, 2015(7):194-197. |
| [12] | 任奎, 唐宇, 范昱, 等. 中国西部六省(区)荞麦属稀有种质资源收集与分类鉴定[J]. 植物遗传资源学报, 2021, 22(4):963-970. |
| [13] |
ZHANG L, MA M, LIU L. Identification of genetic locus underlying easy dehulling in rice-tartary for easy postharvest processing of tartary buckwheat[J]. Genes, 2020, 11(4):459-472.
doi: 10.3390/genes11040459 URL |
| [14] |
LIU M, FU Q, MA Z, et al. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum)[J]. Planta, 2019, 249(5):1301-1318.
doi: 10.1007/s00425-019-03089-3 URL |
| [15] |
WANG Y J. CAMPBELL C G. Tartary buckwheat breeding (Fagopyrum tataricum L. Gaertn.) through hybridization with its rice-tartary type[J]. Euphytica, 2007, 156(3):399-405.
doi: 10.1007/s10681-007-9389-3 URL |
| [16] |
QIAO Z, DIXN R A. Transcriptional networks for lignin biosynthesis: more complex than we thought[J]. Trends in plant science, 2011, 16(4):227-233.
doi: 10.1016/j.tplants.2010.12.005 URL |
| [17] | CHRISTIANSEN L C, DEGAN F D, ULVSKOV P, et al. Examination of the dehiscence zone in soybean pods and isolation of a dehiscence-related endopolygalacturonase gene[J]. Plant cell & environment, 2010, 25(4):479-490. |
| [18] |
ZHANG Q, TU B, LIU C, et al. Pod anatomy, morphology and dehiscing forces in pod dehiscence of soybean (Glycine max (L.) Merrill)[J]. Flora, 2018, 248:48-53.
doi: 10.1016/j.flora.2018.08.014 URL |
| [19] | JOSEFSSON E. Investigations into shattering resistance of cruciferous oil crops[J]. Zeitschrift fur pflanzenzuchtung, 1968, 59:384-96. |
| [20] |
LOLLE S J, PRUITT R E. Epidermal cell interactions: a case for local talk[J]. Trends in plant science, 1999, 4:14-20.
doi: 10.1016/S1360-1385(98)01353-3 URL |
| [21] | HERNANDEZ L F, BELLES P M, BIDEGAIN M A, et al. Biomechanical proposal as a cause of incomplete seed and pericarp development of the sunflower (Helianthus annuus L.) fruits[J]. Phyton international journal of experimental botany, 2018, 87:198-208. |
| [22] | 张超, 李冀新. 荞麦壳的研究进展[J]. 粮油食品科技, 2006(3):8-9. |
| [23] |
ZHU F M, DU B, LI R F, et al. Effect of micronization technology on physicochemical andantioxidant properties of dietary fiber from buckwheat hulls[J]. Biocatalysis and agricultural biotechnology, 2014, 3(3):30-34.
doi: 10.1016/j.bcab.2013.12.009 URL |
| [24] |
CHABANNES M, RUEL K, YOSHINAGA A, et al. In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels[J]. Plant journal, 2001, 28:271-282.
doi: 10.1046/j.1365-313X.2001.01159.x URL |
| [25] |
JONES L, ENNOS A R, TURNER S R. Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis[J]. Plant journal, 2001, 26:205-216.
doi: 10.1046/j.1365-313x.2001.01021.x URL |
| [26] |
VANHOLME R, DEMEDTS B, MORREEL K, et al. Lignin biosynthesis and structure[J]. Plant physiology, 2010, 153:895-905.
doi: 10.1104/pp.110.155119 URL |
| [27] |
PENG D, CHEN X, YIN Y, et al. Lodging resistance of winter wheat (Triticum aestivum L.): lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid[J]. Field crops research, 2014, 157:1-7.
doi: 10.1016/j.fcr.2013.11.015 URL |
| [28] |
WANG C, HU D, LIU X, et al. Effects of uniconazole on the lignin metabolism and lodging resistance of culm in common buckwheat (Fagopyrum esculentum M.)[J]. Field crops research, 2015, 180:46-53.
doi: 10.1016/j.fcr.2015.05.009 URL |
| [29] | 吴朝昕. 薄壳苦荞果壳结构及其发育中的转录组学分析[D]. 贵阳: 贵州师范大学, 2020. |
| [30] |
SONG C, MA C, XIANG D. Variations in accumulation of lignin and cellulose and metabolic changes in seed hull provide insight into dehulling characteristic of tartary buckwheat seeds[J]. International journal of molecular sciences, 2019, 20(3):524-530.
doi: 10.3390/ijms20030524 URL |
| [31] |
SUANUM W, SOMTA P, KONGJAIMUN A, et al. Colocalization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea[J]. Molecular breeding, 2016, 36:80-93.
doi: 10.1007/s11032-016-0505-8 URL |
| [32] | 李淑久, 张惠珍, 袁庆军. 四种荞麦生殖器官的形态学研究[J]. 贵州农业科学, 1992(6):32-36. |
| [33] |
LI H, WU C, LV Q, et al. Comparative cellular, physiological and transcriptome analyses reveal the potential easy dehulling mechanism of rice-tartary buckwheat (Fagopyrum Tararicum)[J]. BMC plant biology, 2020, 20(1):505.
doi: 10.1186/s12870-020-02715-7 URL |
| [34] | 吕文彦, 邹清敏, 郭玉华, 等. 稻谷颖壳开裂特点及其影响的初步研究[J]. 吉林农业大学学报, 2003(3):246-249. |
| [35] |
PATTERSON S E. Cutting loose abscission and dehiscence in Arabidopsis[J]. Plant physiol. 2001, 126(2):494-500.
doi: 10.1104/pp.126.2.494 URL |
| [36] | ESTORNELL L H, AGUSTI J, MERELO P, et al. Elucidating mechanisms underlying organ abscission[J]. Plant science, 2013,199-200. |
| [37] |
CAI S, LASHBROOK C C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in trans-genic plants overexpressing Arabidopsis zinc finger protein2[J]. Plant physiol., 2008, 146(3):1305-13121.
doi: 10.1104/pp.107.110908 URL |
| [38] |
OGAWA M, KAY P, WILSON S, et al. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and quartet2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis[J]. Plant cell, 2009, 21(1):216-233.
doi: 10.1105/tpc.108.063768 URL |
| [39] | SEXTON R, ROBERTS J A. Cell biology of abscission[J]. Annual review of plant physiology, 1982:33-38. |
| [40] | CRISTINA F. Regulation of fruit dehiscence in Arabidopsis[J]. Journal of experimental botany, 2002, 377:2031-2039. |
| [41] | TAYLOR J E, TUCKER G A, LASSLETT Y, et al. Polygalacturonase expression during leaf abscission of normal and transgenic tomato plants[J]. Planta, 1991, 183:133-138. |
| [42] | MEAKIN P J, ROBERTS J A. Dehiscence of fruit in oilseed rape (Brassica napus L.)[J]. Journal of experimental botany, 2008, 8:995-1002. |
| [43] | ULVSKOV P, CHILD R, ONCKELEN H V, et al. Seed shattering, US06420628B1[P]. 2002. |
| [44] | LUNDQVIST U, FRANCKOWIACK J D. Diversity of barley mutants[J]. Developments in plant genetics and breeding, 2003, 7(3):77-96. |
| [45] | KIM J, SUNDARESAN S, PHILOSOPH H S, et al. Examination of the abscission-associated transcriptomes for soybean, tomato, and Arabidopsis highlights the conserved biosynthesis of an extensible extracellular matrix and boundary layer[J]. Frontiers in plant pcience, 2015, 6:1109-1118. |
| [46] | SMIRNOVA A, LEIDE J, RIEDERER M. Deficiency in a very-long-chain fatty acid beta-ketoacyl-coenzyme a synthase of tomata impairs microgametogenesis and causes floral organ fusion[J]. Plant physiol., 2013, 161(1):196-209. |
| [47] |
LOLLE S J, PRUITT R E. Epidermal cell interactions: a case for local talk[J]. Trends in plant science, 1999, 4:14-20.
doi: 10.1016/S1360-1385(98)01353-3 URL |
| [48] |
SIEBER P, SCHODERET M, RYSER U, et al. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions[J]. Plant cell, 2000, 12:721-38.
doi: 10.1105/tpc.12.5.721 URL |
| [49] | TAKETA S, AMANO S, TSUJINO Y, et al. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway[J]. Proceedings of the national academy of sciences of the united states of america, 2008, 105(10):4062-4067. |
| [50] |
KAKEDA K, ISHIHARA N, IZUMI Y, et al. Expression and functional analysis of the barley Nud gene using transgenic rice[J]. Breeding science, 2011, 61:35-42.
doi: 10.1270/jsbbs.61.35 URL |
| [51] |
AHARONI A, DIXIT S, JETTER R, et al. The shine clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis[J]. Plant cell, 2004, 16(9):2463-80.
doi: 10.1105/tpc.104.022897 URL |
| [52] | BROUN P, POINDEXTER P, OSBORNE E, et al. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis[J]. Proceedings of the national academy of sciences of the United States of America, 2004, 101(13):4706-4711. |
| [53] |
KANNANGARA R, BRANIGAN C, LIU Y, et al. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana[J]. Plant cell, 2007, 19(4): 1278-1294.
doi: 10.1105/tpc.106.047076 URL |
| [54] |
RIECHMANN J L, RATCLIFFE O J. A genomic perspective on plant transcription factors[J]. Current opinion in plant biology, 2000, 3:423-434.
doi: 10.1016/S1369-5266(00)00107-2 URL |
| [55] | 周善滋. 油菜等作物的花药开裂结构机理[J]. 湖南师范大学自然科学学报, 1984(1):67-76. |
| [56] |
KIM H J, TRIPLETT B A. Cotton fiber growth in planta and in vitro. models for plant cell elongation and cell wall biogenesis[J]. Plant physiology, 2001, 127(4):1361-1366.
doi: 10.1104/pp.010724 URL |
| [57] |
张雨, 赵明洁, 张蔚. 植物次生细胞壁生物合成的转录调控网络[J]. 植物学报, 2020, 55(3):351-368.
doi: 10.11983/CBB19135 |
| [58] | PASCUAL M B, TORRE F, CAAS R A, et al. NAC transcription factors in woody plants[J]. Progress in botany, 2018, 80:195-222. |
| [59] |
AIDA M, ISHIDA T, FUKAKI H, et al. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant[J]. Plant cell, 1997, 9(6):841-57.
doi: 10.1105/tpc.9.6.841 URL |
| [60] |
SOUER E, VAN H A, KLOOS D, et al. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries[J]. Cell, 1996, 85(2):159-170.
doi: 10.1016/S0092-8674(00)81093-4 URL |
| [61] |
MITSUDA N, SEKI M, SHINOZAKI K, et al. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence[J]. Plant cell, 2005, 17(11):2993-3006.
doi: 10.1105/tpc.105.036004 URL |
| [62] |
MITSUDA N, MASARU O T. NAC Transcription Factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis[J]. Plant cell, 2007, 19(1):270-280.
doi: 10.1105/tpc.106.047043 URL |
| [63] | ZHONG R, YE Z H. The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems[J]. Plant signaling, behavior, 2015, 10(2):989746. |
| [64] |
WANG J, MA Z T, TANG B, et al. Tartary Buckwheat (Fagopyrum tataricum) NAC transcription factors FtNAC16 negatively regulates of pod cracking and salinity tolerant in Arabidopsis[J]. International journal of molecular sciences, 2021, 22(6):3197-3203.
doi: 10.3390/ijms22063197 URL |
| [65] |
SCHIEFELBEIN J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot[J]. Current opinion in plant biology, 2003, 6(1):74-78.
doi: 10.1016/S136952660200002X URL |
| [66] |
TOLEDO O G, QUAIL H P. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. Plant cell, 2003, 15(8):1749-1770.
doi: 10.1105/tpc.013839 URL |
| [67] |
DUEK P D, FANKHAUSER C. bHLH class transcription factors take centre stage in phytochrome signalling[J]. Trends in plant science, 2005, 10:51-54.
doi: 10.1016/j.tplants.2004.12.005 URL |
| [68] | KIM J, KIM H Y. Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response[J]. BBA-gene structure and expression, 2006, 1759(3):191-194. |
| [69] |
HESLOP H J. Pollen wall development. the succession of events in the growth of intricately patterned pollen walls is described and discussed[J]. Science, 1968, 161(3838):230-237.
doi: 10.1126/science.161.3838.230 URL |
| [70] | 宋垚, 李晖, 石其龙, 等. 拟南芥bHLH家族转录因子DYT1在花药发育过程中调控胼胝质降解[J]. 上海师范大学学报, 2009, 38(2):174-182. |
| [71] |
RAJANI S, SUNDARESAN V. The Arabidopsis myc/bHLH gene alcatraz enables cell separation in fruit dehiscence[J]. Current biology, 2001, 11:1914-1922.
doi: 10.1016/S0960-9822(01)00593-0 URL |
| [72] |
LILJEGREN S J, ROEDER A, KEMPIN S, et al. Control of fruit patterning in Arabidopsis by indehiscent[J]. Cell, 2004, 116:843-853.
doi: 10.1016/S0092-8674(04)00217-X URL |
| [73] |
SUN W J, JIN X, MA Z T, et al. Basic helix-loop-helix (bHLH) gene family in tartary buckwheat (Fagopyrum tataricum): genome-wide identification, phylogeny, evolutionary expansion and expression analyses[J]. International journal of biological macromolecules, 2020, 155:1478-1490.
doi: 10.1016/j.ijbiomac.2019.11.126 URL |
| [74] |
MASAKI F, TOSHITSUGU N, YOKO S, et al. A large-scale identification of direct targets of the tomato MADS box transcription factor ripening inhibitor reveals the regulation of fruit ripening[J]. Plant cell, 2013, 25:371-386.
doi: 10.1105/tpc.112.108118 URL |
| [75] |
GU Q, FERRANDIZ C, YANOFSKY M F, et al. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development[J]. Development, 1998, 125(8):1509-1517.
doi: 10.1242/dev.125.8.1509 URL |
| [76] |
LILJEGREN S J, DITTA G S, ESHED Y, et al. Shattesrproof MADS-box genes control seed dispersal in Arabidopsis[J]. Nature, 2000, 404:766-770.
doi: 10.1038/35008089 URL |
| [77] |
FERRANDIZ C, LILJEGREN S J, YANOFSKY M F. Negative regulation of the shatterproof genes by fruitfull during Arabidopsis fruit development[J]. Science, 2000, 289(5478):436-438.
doi: 10.1126/science.289.5478.436 URL |
| [78] |
MA D, CONSTABEL C P. MYB repressors as regulators of phenylpropanoid metabolism in plants[J]. Trends in plant science, 2019, 24(3):275-289.
doi: 10.1016/j.tplants.2018.12.003 URL |
| [79] | 朱晓博, 张贵粉, 陈鹏. 植物次生细胞壁加厚过程的转录调控[J]. 植物生理学报, 2017, 53(9):1598-1608. |
| [80] |
RAMSAY N A, GLOVER B J. MYB-bHLH-WD 40 protein complex and the evolution of cellular diversity[J]. Trends in plant science, 2005, 10:63-70.
doi: 10.1016/j.tplants.2004.12.011 URL |
| [81] | LI C, NG C K, FAN L M, et al. MYB transcription factors, active players in abiotic stress signaling[J]. Environmental andexperimental botany, 2015, 114: 80-91. |
| [82] | ROY S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome[J]. Plant signaling behavior, 2016, 11:1-7. |
| [83] |
LIU J Y, OSBOURN A, MA P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants[J]. Molecular plant, 2015, 8:689-708.
doi: 10.1016/j.molp.2015.03.012 URL |
| [84] |
SEO M S, KIM J S. Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae[J]. Molecules, 2017, 22:1-13.
doi: 10.3390/molecules22010001 URL |
| [85] |
ZHONG R, RICHARDSON E A, YE Z H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis[J]. Plant cell, 2007, 19(9):2776-2792.
doi: 10.1105/tpc.107.053678 URL |
| [86] |
ZHONG R, LEE C, ZHOU J, et al. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis[J]. Plant cell, 2008, 20(10):2763-2782.
doi: 10.1105/tpc.108.061325 URL |
| [87] |
STEINER L S, UNTE U S, ECKSTEIN L, et al. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers[J]. Plant journal, 2003, 34(4):519-528.
doi: 10.1046/j.1365-313X.2003.01745.x URL |
| [88] |
YANG C, XU Z, SONG J, et al. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence[J]. The plant cell online, 2007, 19(2):534-548.
doi: 10.1105/tpc.106.046391 URL |
| [89] | YANG J B, SOMERS D A, WRIGHT R L, et al. Seed pod dehiscence in birdsfoot trefoil, lotus conimbricensis, and their interspecific somatic hybrid[J]. Canadian journal of plant, 1990, 70(1):279-284. |
| [1] | 巩永永, 端木慧子. 甜菜TIFY基因家族的全基因组鉴定与生物信息学分析[J]. 中国农学通报, 2022, 38(8): 17-24. |
| [2] | 余兰, 王浩然, 张莹, 邢红运, 丁琪, 赵宝珍, 崔娜. 转录因子MYCs调控番茄表皮毛萜类化合物的分子机制研究进展[J]. 中国农学通报, 2022, 38(6): 87-93. |
| [3] | 徐晓美, 李颖, 衡周, 徐小万, 李涛, 王恒明. 响应辣椒疫霉菌诱导的CaWRKY转录因子筛选及其信号通路分析[J]. 中国农学通报, 2022, 38(32): 22-31. |
| [4] | 马慧敏, 孙培琳, 马春泉. 转录因子BvM14-GAI耐盐功能研究[J]. 中国农学通报, 2021, 37(34): 34-42. |
| [5] | 王雪, 王盛昊, 于冰. 转录因子和启动子互作分析技术及其在植物应答逆境胁迫中的研究进展[J]. 中国农学通报, 2021, 37(33): 112-119. |
| [6] | 张晋, 刘思辰, 曹晓宁. 不同荞麦品种种壳营养成分研究[J]. 中国农学通报, 2021, 37(32): 132-138. |
| [7] | 孙铭阳, 徐世强, 顾艳, 梅瑜, 周芳, 李静宇, 王继华. 穿心莲全长转录组测序及特性分析[J]. 中国农学通报, 2021, 37(27): 82-89. |
| [8] | 张璐, 何录秋, 杨学乐. 不同镉背景值农田中荞麦镉积累转运特性研究[J]. 中国农学通报, 2021, 37(23): 77-83. |
| [9] | 杜晓雪, 黄园园, 马春泉, 李海英. 转录因子BvM14-Dof3.4响应盐胁迫的功能研究[J]. 中国农学通报, 2021, 37(21): 119-125. |
| [10] | 刘恺媛, 王茂良, 辛海波, 张华, 丛日晨, 黄大庄. 植物花青素合成与调控研究进展[J]. 中国农学通报, 2021, 37(14): 41-51. |
| [11] | 李英, 杜春梅. 致病性尖孢镰刀菌毒力因子的研究进展[J]. 中国农学通报, 2021, 37(12): 92-97. |
| [12] | 尹桂芳, 李春花, 孙道旺, 卢文洁, 王艳青, 王莉花. 苦荞麦不同部位干燥后DNA提取效果的比较及薄壳性状关联SSR引物筛选研究[J]. 中国农学通报, 2020, 36(6): 69-73. |
| [13] | 谭景发, 贺文闯, 董西龙, 党腾飞, 谢怿, 席锟, 孙勇胜, 胡亚林, 靳德明. 不同水稻种质中渗透胁迫抗性基因DREB2A的遗传多样性分析[J]. 中国农学通报, 2020, 36(35): 1-13. |
| [14] | 于冰,田烨,李海英,吕笑言,王宇光,端木慧子. 植物bHLH转录因子的研究进展[J]. 中国农学通报, 2019, 35(9): 75-80. |
| [15] | 朱长保,徐辰峰,刘仁梅. 干旱胁迫下水稻转录因子表达变化[J]. 中国农学通报, 2019, 35(6): 108-114. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||